MGEScan-nonLTR¶
MGEScan-nonLTR is a program to identify non-long terminal repeat (non-LTR) retrotransposons in genomic sequences. A few options are available in the Galaxy workflow system to configure the program settings, e.g. hmmsearch of protein sequence database with a profile hidden Markov model (HMM).
Description¶
MGEScan-nonLTR identifies non-LTR retrotransposons based on Gaussian Bayes classifiers and generalized hidden Markov models consisting of twelve super states that correspond to different clades or closely related clades.
For details, please read following reference.
- Rho, M., Tang, H. (2009) MGEScan-non-LTR: computational identification and classification of autonomous non-LTR retrotransposons in eukaryotic genomes. Nucleic Acids Research, 37(21), e143.
Running the program¶
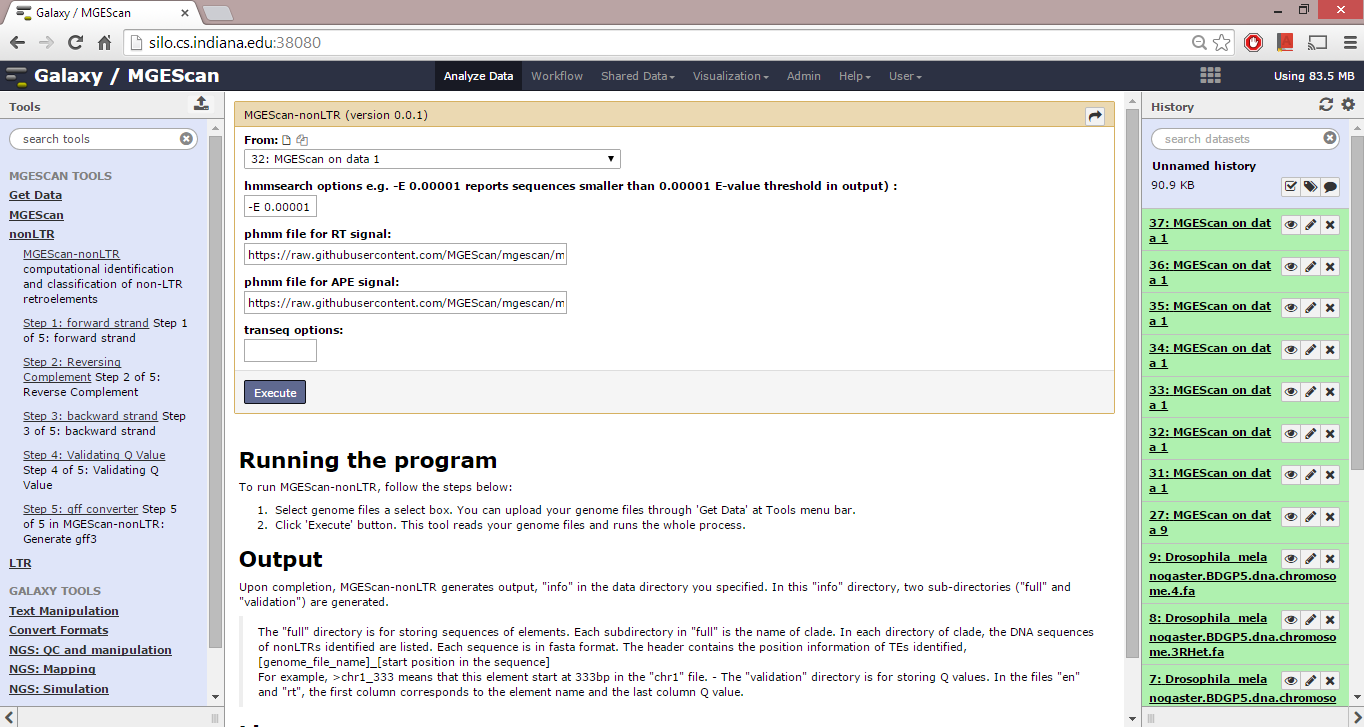
To run MGEScan-nonLTR, follow the steps below:
- Select genome files a select box. You can upload your genome files through ‘Get Data’ at Tools menu bar.
- Click ‘Execute’ button. This tool reads your genome files and runs the whole process.
Options¶
- hmmmsearch options e.g. -E 0.00001 : reports sequences smaller than 0.00001 E-value threshold in output
- URL of the profile files for RT and APE
- EMBOSS transeq options
Results¶
Upon completion, MGEScan-nonLTR generates output, “info” in the data directory you specified. In this “info” directory, two sub-directories (“full” and “validation”) are generated.
The “full” directory is for storing sequences of elements. Each subdirectory in “full” is the name of clade. In each directory of clade, the DNA sequences of nonLTRs identified are listed. Each sequence is in fasta format. The header contains the position information of TEs identified, [genome_file_name]_[start position in the sequence] For example, >chr1_333 means that this element start at 333bp in the “chr1” file. - The “validation” directory is for storing Q values. In the files “en” and “rt”, the first column corresponds to the element name and the last column Q value.
License¶
Copyright 2015. You may redistribute this software under the terms of the GNU General Public License.