QuickStart¶
MGEScan, identifying LTR and non-LTR in genome sequences are available on the Galaxy scientific workflow which is a web-based workflow software to support data analysis with various tools.
Overview¶
This tutorial demonstrates a quick start of using MGEScan on Galaxy workflow with a sample dataset, D. melanogaster genome.
Tip
Approximate 3 hours and 30 minutes (including 3 hours computation time)
Run MGEScan-LTR and MGEScan-nonLTR for D. melanogaster¶
In this tutorial, we will try to run both MGEScan-LTR and MGEScan-nonLTR with
D. melanogaster genome dataset. You can find the dataset at the Shared Data
menu on top and MGEScan tools on the left frame.
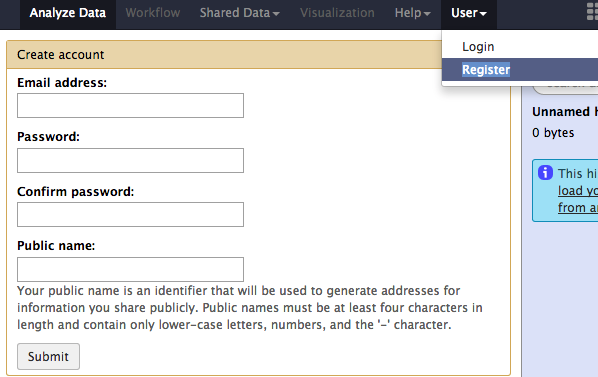
Login or Register (Optional)¶
You can save your work if you have account on Galaxy workflow. The user-based history in Galaxy/MGEScan stores your data and launched tasks. The guest user account is able to run the MGEScan tools without the login but results or history data won’t be saved if the web browser session is closed.
Login¶
If you already have an account, you can use your user id and password at the User > Login page.
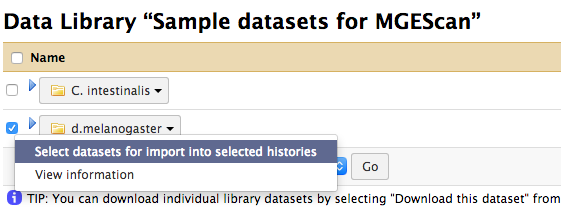
Example: Drosophila melanogaster¶
In the Data Library, enable the checkbox for d.melanogaster and click
“Select datasets for import into selected histories” from the down arrow at
the end.
You will find 8 fasta files are available. We need to import all of them, make them all checked and click “Import library datasets” in the middle of the page.

Once you imported the D. melanogaster datasets into your history, you are ready to run MGEScan tools on Galaxy. Go to the main page, and checkout imported datasets (8 files) on the right frame of the page.
Note
You can select where datasets to be imported.
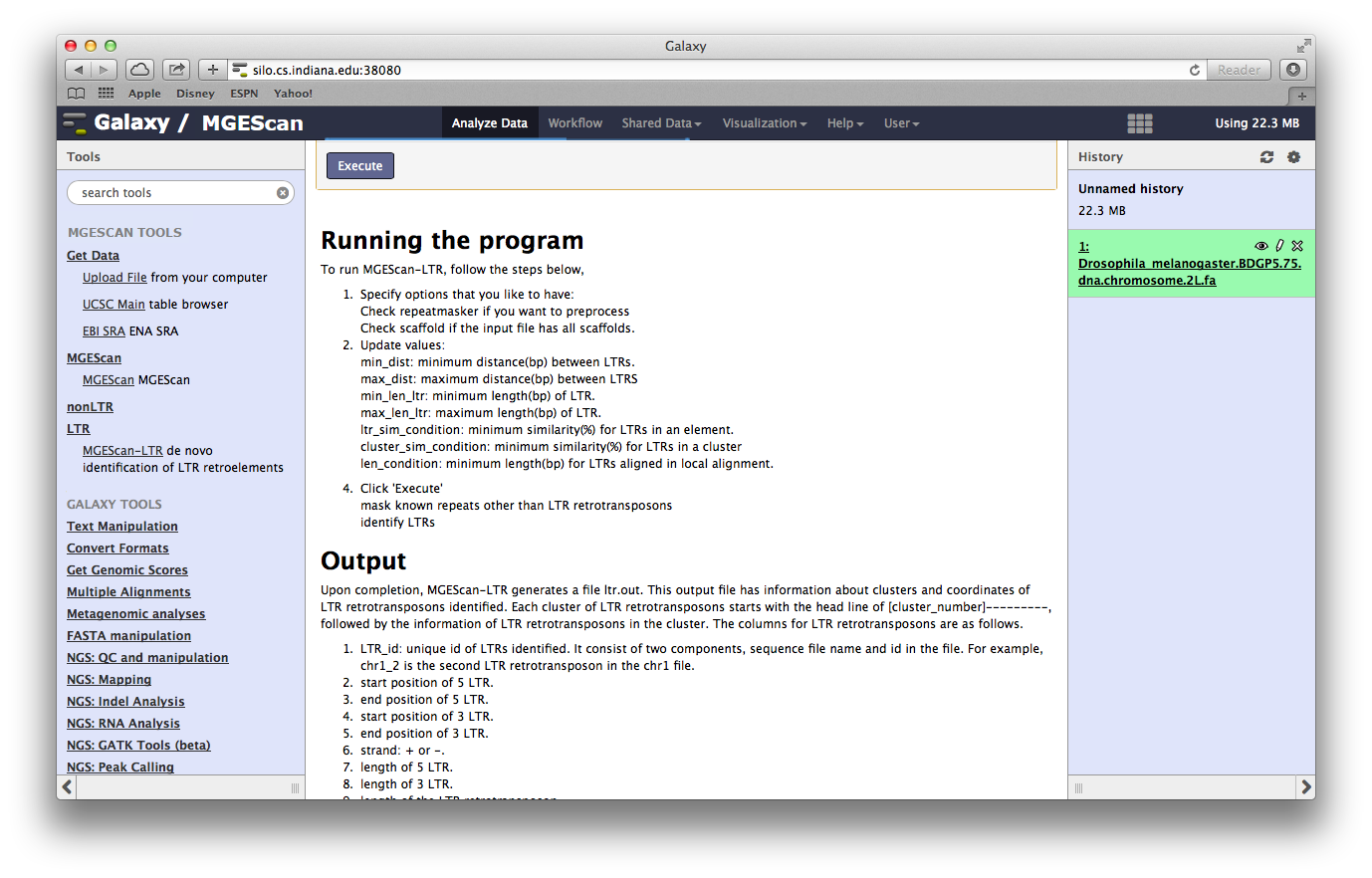
Run MGEScan for LTR and nonLTR¶
In the new version of MGEScan, two programs, MGEScan-LTR and MGEScan-nonLTR, can be ran at the same time with a merged result. Open the page at “MGEScan > MGEScan”, a simple tool is available for LTR and nonLTR executions with MPI option for parallel processing.
Note
Find LTR or nonLTR page if you’d like to choose other options to run MGEScan tools in detail.
Create a single link to multiple inputs¶
In the example of d. melanogaster, we have 8 fasta files as its sequences.
To analyze them all at the same time, we create a single link to the files
prior to running MGEScan tool on Galaxy. One archive file to many files (e.g.
file.tar) will be used as an input of MGEScan tool on Galaxy. Note that Galaxy
workflow does not support multiple arbitrary inputs but this symlink tool
allows you to have dynamic inputs as a Galaxy input dataset.
- FInd “Tools > Create a symlink to multiple datasets” on the left frame.
We will add 8 fasta files each by clicking “Add new Dataset” from “8: Drosophila_melanogaster.BDGP6.dna.chromosome.dmel_mitochondrion_genome.fa” to “1: Drosophila_melanogaster.BDGP6.dna.chromosome.2L.fa” like so:
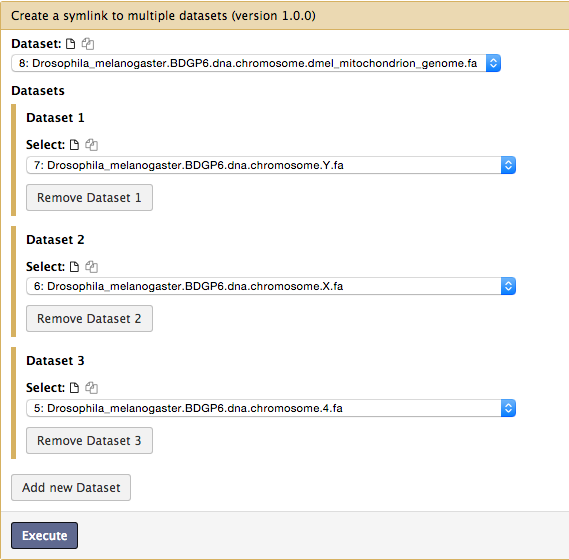
Make sure you have added all the files without duplication. The added order is not important though. File(s) will be placed in a same directory without order.
MGEScan Tool¶
MGEScan runs both LTR and nonLTR with a selected input genome sequence. Find “MGEScan > MGEScan” tool on the left frame and confirm that the symlink dataset we created in the previous step is loaded in “From” select form like so:
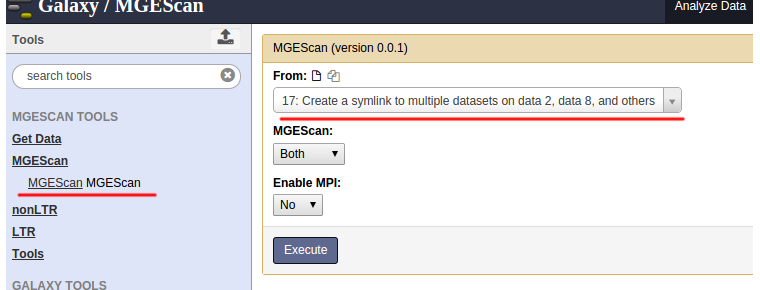
Enable MPI¶
To accelerate processing time, select “Yes” at “Enable MPI” select form and specify “Number of MPI Processes”. If you have a multi-core system, use up to the number of cores.
Our options are:
- From: Create a symlink to multiple datasets on data 2 and data 8, and others
- MGEScan: Both
- Enable MPI: Yes
- Number of MPI Processes: 4
And click “Execute”.
Computation Time¶
Our test case took 3 hours for analyzing LTR and nonLTR of D. melanogaster:
- nonLTR: 19 minutes
- LTR: 3 hours
- Total: 3 hours
Results¶
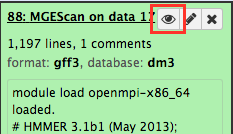
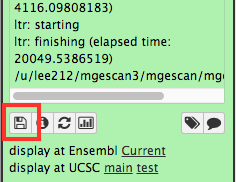
Upon the MGEScan tools completion, the output files are accessible via Galaxy in gff3 format, a plain text, or an archived (e.g. tar.gz) file. You will notice that the color of your tools has been changed to green like so:
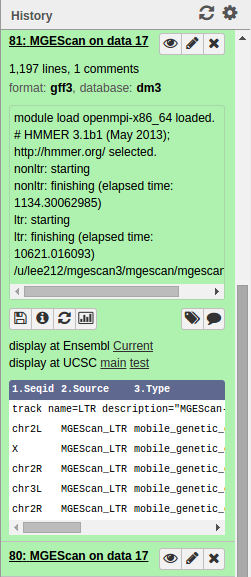
You can download the output files to your local storage, or get access to Genome Browser with provided links.
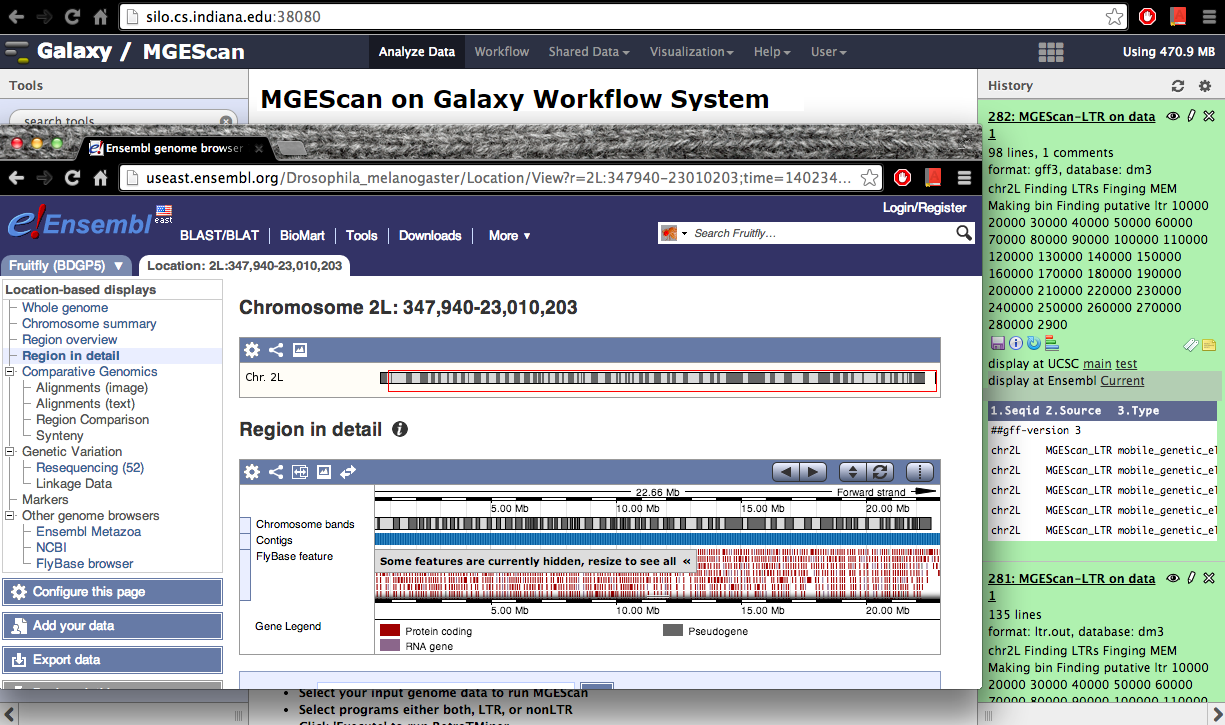
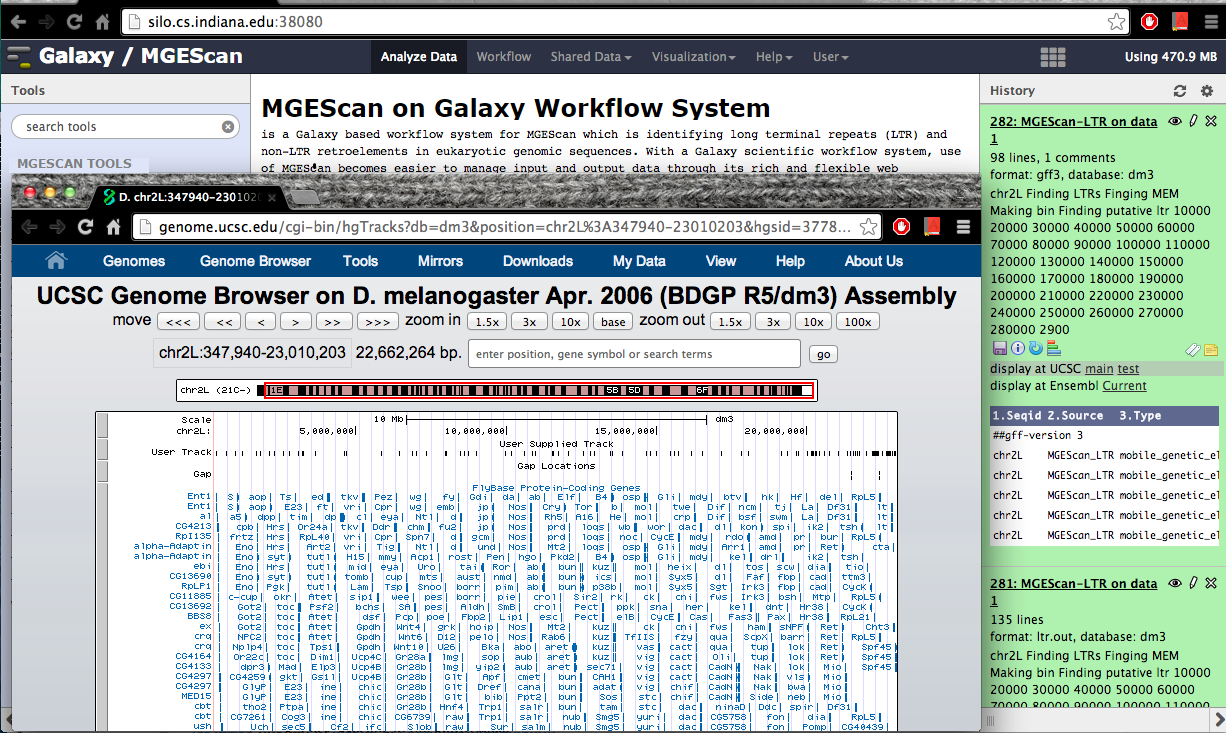
Visualization: UCSC or Ensembl Genome Browser¶
Your genomic data in a Generic Feature Format Version 3 (gff3) can be displayed by a well known visualization tool such as UCSC or Ensembl Genome Browser on Galaxy with custom annotations of MGEScan for LTR and nonLTR. Find the link provided for gff3 to view interactive graphical display of genome sequence data.
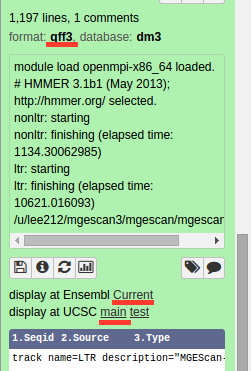
UCSC Genome Browser (Example View)¶
Ensembl (Example View)¶